| Introduction to the visualization program VMD |
|---|
"VMD is a molecular visualization program for displaying, animating, and analyzing large biomolecular systems using 3-D graphics and built-in scripting. VMD supports computers running MacOS-X, Unix, or Windows."
VMD is developped by the Theoretical and Computational Biophysics Group at the National Institute of Health.
User's manual:Here you can find the online HTML version as well as a pdf version (1011 KB) and a PostScript (6.6 MB) version. You may alternatively download the tar/gzip HTML version (383 KB).
For our conveinece we intend to make VMD available on the Unix computers. Alternatively you may install it on our personal computer. For detailed instructions and licensing issues please refer to the VMD site.
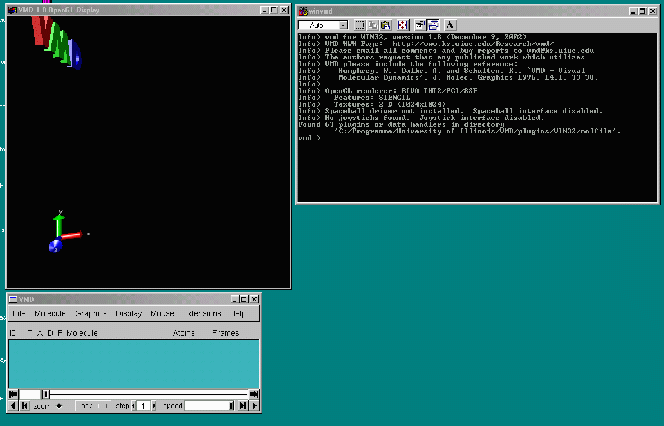
After successfully starting VMD you will find your (PC) screen looking like this:
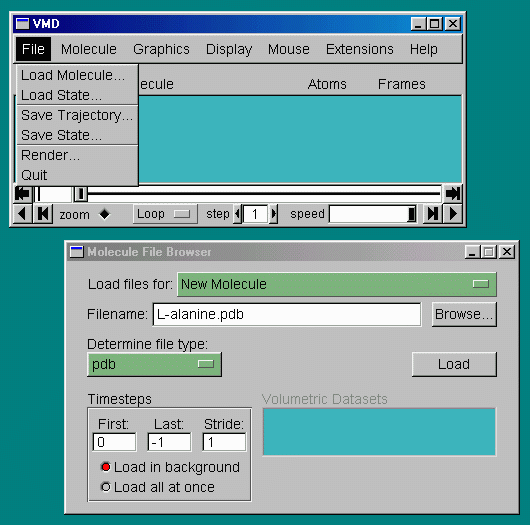
To load to a molecule use the pull-down menu "File" and chose "Load Molecule" which will bring up the Molecule File Browser window:
After you have chosen a suitable molecule (L-alanine) and confirmed the "Load" button, the display window should look like this:
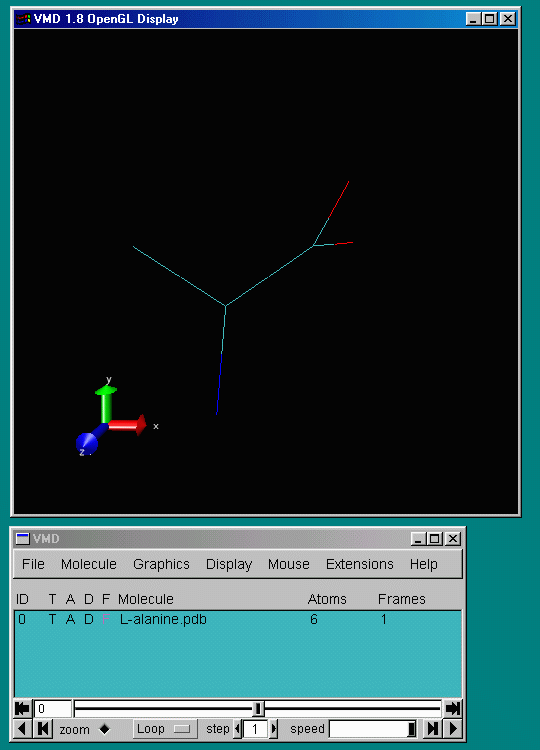
Shown is a wireframe representation of the molecule. At the end
of each line sits an atom.
Carbon is coloured cyan,
nitrogen blue,
oxygen red, and hydrogen white.
Try to write down the chemical structure of this molecule. Why are
there no hydrogens present ?
Hint: Have a look at the L-alanine.pdb file using a text browser.
Now make yourselfs familar how the movements and buttons of your mouse
affect the
orientation of the molecule. You should be able perform rotations and
zoom.
Next step: Graphics and representations