Researcher of this topic is: Barbara Hutter
Genomic Imprinting
Imprinting is a special mechanism of gene regulation in mammals like human, mouse, cattle, and sheep. In human and mouse, approximately 70 imprinted genes are known [1,2]. They are monoallelically expressed depending on the parental origin of the chromosome. For example, the insulin like growth factor gene (Igf2) is only transcribed from the chromosome transmitted by the father, whereas the insulin like growth factor receptor gene (Igf2r) is only expressed on the chromosome inherited from the mother.
A distinction between the paternal and the maternal allele is possible by different epigenetic modifications of the DNA, such as methylation of cytosines in CpG dinucleotides. So-called differentially methylated regions are methylated on one allele but unmethylated on the other. Often they overlap with CpG islands which are associated with the promoter region of genes (Figure 1). If the CpG island is methylated, the chromatin structure of the promoter region is thought to become dense, causing transcriptional silencing of the associated gene.
Imprinted genes play important roles in embryonic development. Proteins encoded by them take part in many pathways and interactions, including transcription factors involved in regulatory cascades. In the light of these functions, it is evident that imprinting disorders, which cause either over- or underexpression of the genes, result in severe diseases like growth anomalies, behaviour anomalies, and cancer.
As a collaboration between the Chair for Computational Biology and Dr. Martina Paulsen from the Chair for Genetics of the Saarland University, our research is concentrated on DNA sequence analysis of mammalian imprinted genes and their genomic environment with bioinformatics methods. Repetitive elements show a specific distribution around imprinted genes [3]. We found that the CpG islands of imprinted human and mouse genes are enriched in tandem repeats [4]. There is, however, little sequence conservation of these tandem repeats. The special chromatin structure provided by CpG rich and/or repetitive sequences seems to be of particular relevance. Moreover, CpG islands show different features between the two species. This is a general issue that we recently adressed in a related study using different CpG island identification programs [5,6]. We could confirm earlier reports that human CpG islands are longer and G+C-richer than mouse ones.
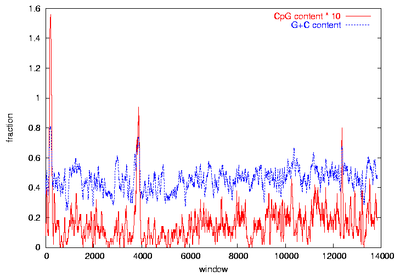
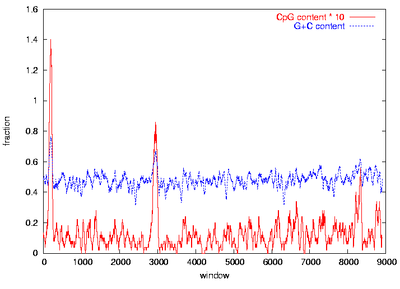
Figure 1: Visual CpG island identification
CpG plots of the insulin like growth factor receptor genes from human (IGF2R, right) and mouse (Igf2r, left) were created by calculation of G+C and CpG content in sliding windows of 500 bp moved by 10 bp. CpG islands can be assigned visually as peaks of the CpG content (red) with at least 6 % CpG where also the G+C content (blue) exceeds 60 %. Thus, a prominent CpG island can be identified in the promoter region around the gene start, and two weaker ones are located inside of the gene.
Comparative genomics
Our current project “Sequenzbasierte Analyse der Regulation und Interaktion von imprinteten Genen” is funded by the DFG (PA 750/3-1). Here, we focus on comparative genomics and phylogenetic footprinting to detect evolutionary conserved motifs also outside of CpG islands in a larger set of orthologous imprinted sequences. Evolutionary conserved regions provide promising candidates for regulatory elements whose function can be examined in the laboratory. When located around the start of a gene, i.e. in the promoter region, they may harbor transcription factor binding sites (Figure 2).

Figure 2: Conserved potential transcription factor binding sites
Using web-based comparative genomics tools from DCODE.org [7] on the promoter CpG island sequences of human IGF2R and mouse Igf2r, a number of conserved potential transcription factor binding sites could be identified. KROX, EGR, and STAT factors are involved in growth regulation. They may contribute to a regulatory module for the expression of the insulin like growth factor receptor gene.
Although there exist several tools for comparative genomics, they are limited to pairwise sequence comparisons. Therefore, one of our aims is to develop a strategy to gain multiple conservation information by projecting pairwise conservation to the human respectively mouse sequence, which serve as references.
We will also address the issue of tissue-specific expression of imprinted genes. We hope to distinguish possible imprinting-specific motifs from those responsible for tissue-specific expression. Further research is directed towards the interaction networks between proteins coded by imprinted genes and normal, biallelically expressed genes.
References
[1] Imprinted Gene Catalogue at the University of Otago: http://igc.otago.ac.nz/home.html
[2] Mouse Imprinting at Harwell Mammalian Genetics Unit: http://www.mgu.har.mrc.ac.uk/research/imprinting/index.html
[3] J. Walter, B. Hutter, T. Khare, and M. Paulsen (2006) “Repetitive elements in imprinted genes”, Cytogenet. Genome Res. 113(1-4): 109-115 [abstract]
[4] B. Hutter, V. Helms, and M. Paulsen (2006) “Tandem repeats in the CpG islands of imprinted genes”, Genomics 88(3): 323-332 [abstract]
[5] CpG Island Searcher: http://cpgislands.usc.edu/
[6] Recursive Segmentation of DNA Sequences: http://www.nslij-genetics.org/wli/dnaseg/
[7] DCODE.org Comparative Genomics Center: http://www.dcode.org/