NEW
PROPORES was recently updated and is now available as the standalone tool ProPores2 (github.com) and as a corresponding webservice.
Hollander, M., Rasp, D., Aziz, M., Helms, V. (2021) Journal of Chemical Information and Modeling, doi:10.1021/acs.jcim.1c00154. ProPores2: web service and stand-alone tool for identifying, manipulating, and visualizing pores in protein structures.
Publication
Lee, P.-H. and Helms, V. (2012) Proteins: Structure, Function, and Bioinformatics, vol. 80, p. 421-432, doi: 10.1002/prot.23204. Identifying continuous pores in protein structures with PROPORES by computational repositioning of gating residues.
Introduction
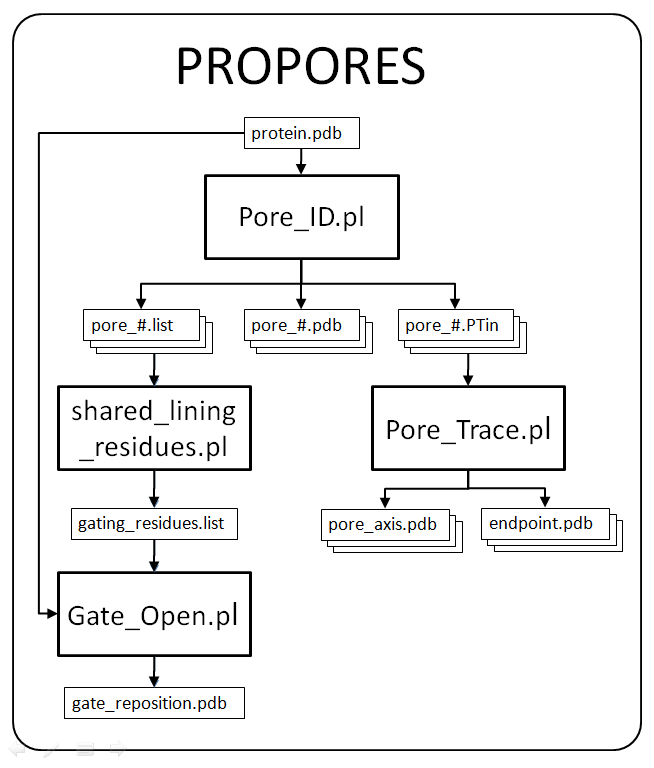
“PROPORES” is a novel toolkit for identifying pockets, cavities and channels of protein structures. The toolkit was developed in PERL programming language and includes “PoreID” for pore identification, “PoreTrace” for pore axes determination and “GateOpen” for opening the gate between neighboring pores. “PoreID” is a grid-based method that avoids orientation dependency of the results. It targets all kinds of pores (pockets, cavities and channels) and is automatic so that only the PDB file of the target protein has to be specified by the user. Several descriptors of pores such as volume, lining residues, axes, and radius profiles are provided by “PoreID” and “PoreTrace”. In addition, for two neighboring pores, “GateOpen” can be used to test connectivity by rotating side chains of the gating residues. This was done to introduce a dynamic variable into the pore identification of the otherwise static protein structure.
Scheme of the toolkit
Download
Here you can download the PROPORES software.
Contact
Po-Hsien Lee
p.lee@mx.uni-saarland.de