Summary
TopoView is a Topology Viewer for alpha-helical transmembrane proteins.
Researcher
Nadine Schneider
Description
Residue-based diagrams of proteins, also called ‘snake plots’, are often employed for representing alpha-helical transmembrane proteins as long as their three-dimensional structures are not available. This 2D representation allows visualizing the helical portions of the protein sequence and, in the case of transmembrane proteins, the topology (i.e. the orientation of the N-terminus with regard to the lipid bilayer). The tool TopoView is such a topology viewer for alpha-helical TM proteins. Given an amino acid sequence and the borders of the TM segments, it produces a high quality 2D representation of this protein sequence. As special features, one may visualize user-specified structural features such as interface helices, disrupted transmembrane helices or non contacting consecutive transmembrane helices.
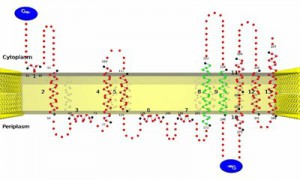
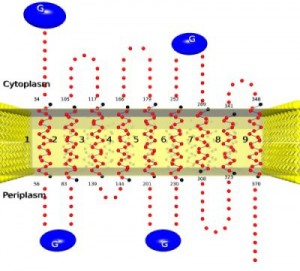
Implementation
The tool TopoView was implemented in Python 2.7 as Python is widespread, platform independent and efficient compilers are freely available. Also, a graphical user interface was integrated. The high-quality images produced by TopoView are rendered by means of the tool POV-Ray (Persistance of Vision Raytracer). POV-Ray is another platform independent open source project that creates stunning three-dimensional graphics. Therefore, current Python and POV-Ray releases are prerequisites for utilizing TopoView.
Installation
The tool TopoView can be downloaded as a stand-alone program for linux systems: TopoView.tar.gz
After downloading the file, unzip the file, go to the folder ‘source’ and then launch the application by executing:
python TopoView.py
The documentation (user’s manual) is located in the folder ‘docs’ of TopoView